https://www.sprpages.nl/data-fitting/theory https://www.sprpages.nl/data-fitting/kinetic-modelss
Data Fitting Theory
Integrated rate equation:
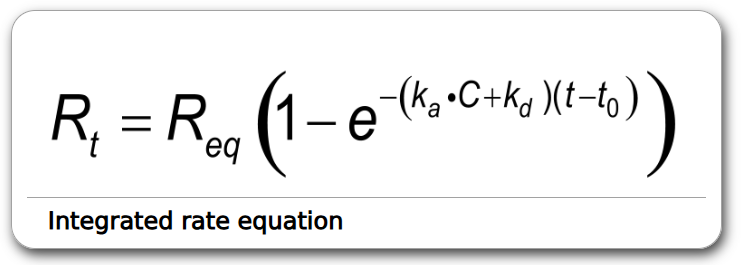
Nonlinear Regression also exists
Assumptions with non-linear regression
The results of non-linear regression are meaningful only if the following assumptions are true (or nearly true):
- The model is correct. Non-linear regression adjusts the variables in the chosen equation to minimize the sum-of squares. It does not attempt to find a better equation.
- The variability of values around the curve follows a Gaussian distribution. Although no biological variable follows a Gaussian distribution exactly, it is sufficient that the variation is approximately Gaussian.
- The SD of the variability is the same everywhere, regardless of the value of X. If the SD is not constant but rather is proportional to the value of Y, the data should be weighted to minimize the sum-of-squares of the relative distances.
- The model assumes that X is known exactly. This is rarely the case, but it is sufficient to assume that any imprecision in measuring X is very small compared to the variability in Y.
- The errors are independent. The deviation of each value from the curve should be random, and should not be correlated with the deviation of the previous or next point.
Kinetic Models
R_max is globally fitted because binding sites may be lost due to regeneration conditions or incomplete regeneration, which makes it unreliable to calculate R_max from a single analyte and ligand concentration/trial
- Globally fit - using the multiple analyte and ligand concentrations
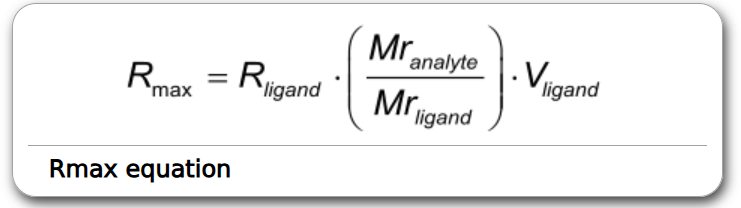
Start with a global fitting of the dissociation rate constant (_k_d). Use the obtained _k_d in the next global fitting with _k_a and Rmax (fix _k_d as a constant). Set the Bulk RI to zero and omit mass transfer and drift. After the fit is made, look at the fitted curves. Are they following the measured data? Is the dissociation fitted correctly? Are the association and Rmax calculated within the expected range?
- What equation do we use to globally fit k_d question
R_max is the maximum signal produced at high analyte concentrations R_eq varies for each analyte concentration

- Are both of the integrated rate equations a part of the software?
- They are both a part of the process, but the PDF does not adequately describe in what order they are used: please see Octet Biomolecular binding kinetics for a proper workflow
Methods for checking validity:
https://www.sprpages.nl/data-fitting/validation
For Km? : Cornish-Bowden, A. - Detection of errors of interpretation in experiments in enzyme kinetics; Methods ;24: 181-190; (2001).